
Digital pathology, is a new, rapidly expanding field in medical imaging in which whole-slide scanners are used to digitise glass slides at high resolution (up to 160nm per pixel). The availability of whole-slide images (WSI) has garnered the interest of the medical image analysis community, resulting in increasing numbers of publications on histopathologic image analysis.
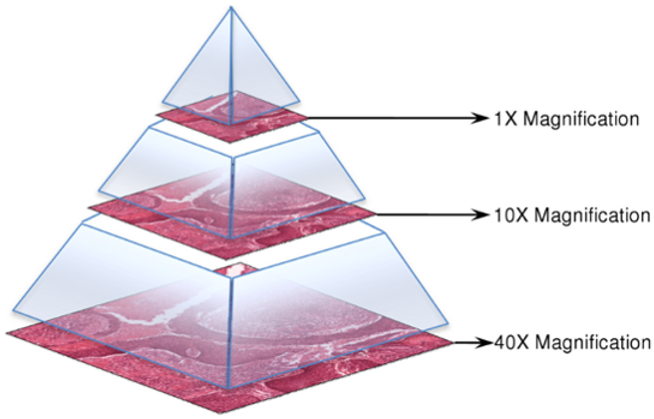
Whole-slide images are generally stored in a multi-resolution pyramid structure. Image files contain multiple down-sampled versions of the original image. Each image in the pyramid is stored as a series of tiles, to facilitate rapid retrieval of subregions of the image.
A typical whole-slide image is approximately 200000 x 100000 pixels on the highest resolution level with 3 byte RGB pixel format. This means 55.88GB of uncompressed pixel data from a single level.
The aim of CAMELYON17 is to evaluate new and existing algorithms for automated detection and classification of breast cancer metastases in whole-slide images of histological lymph node sections. Last year’s CAMELYON16 focused on the detection of lymph node metastases, both on a lesion-level and on a slide-level. This year we move up to patient-level analysis, which requires combining the detection and classification of metastases in multiple lymph node slides into one outcome: a pN-stage. This task has high clinical relevance and would normally require extensive microscopic assessment by pathologists. Therefore, an automated solution for assessing the pN-stage in breast cancer patients, would hold great promise to reduce the workload of pathologists, while at the same time, reduce the subjectivity in diagnosis.