Foundation Model in Medical Image Analysis
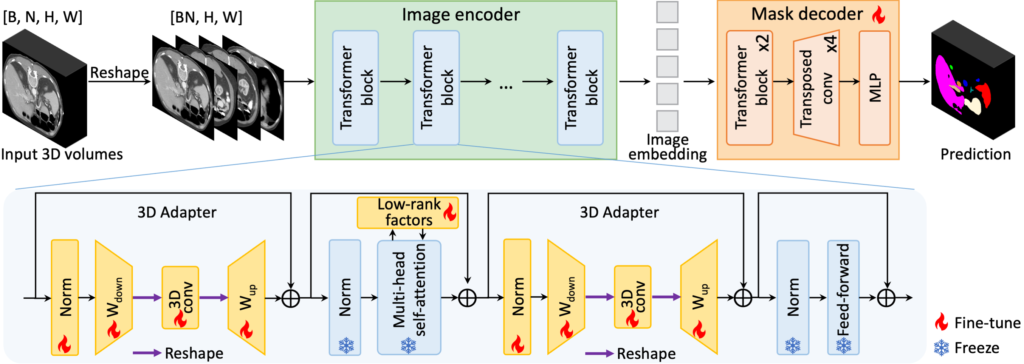
Driven by their remarkable generalization and few-shot learning capability, foundation models have gained significant attention in the field of computer vision. In medical image analysis, there is also rapidly growing interest in adapting pretrained large models to a diversity of downstream tasks, as opposed to the conventional practice of crafting task-specific models from scratch. At CAMCA, we have undertaken broad research in the development of medical imaging foundation models and the adaptation of general visual foundation models to medical applications. Some of our recent projects on medical imaging foundation model include: 1) CMITM, a cross-modal image-text pre-training framework leveraging both masked autoencoding and contrastive learning; 2) MA-SAM, a modality-agnostic SAM adaptation framework for 3D medical image segmentation; 3) MediViSTA-SAM, a spatio-temporal SAM adaptation framework for zero-shot medical video analysis.
Related publications:
CMITM: https://link.springer.com/chapter/10.1007/978-3-031-43904-9_48
MA-SAM: https://arxiv.org/pdf/2309.08842.pdf
MediViSTA-SAM: https://arxiv.org/pdf/2309.13539.pdf
Robust Image Segmentation by Deformable Convolution
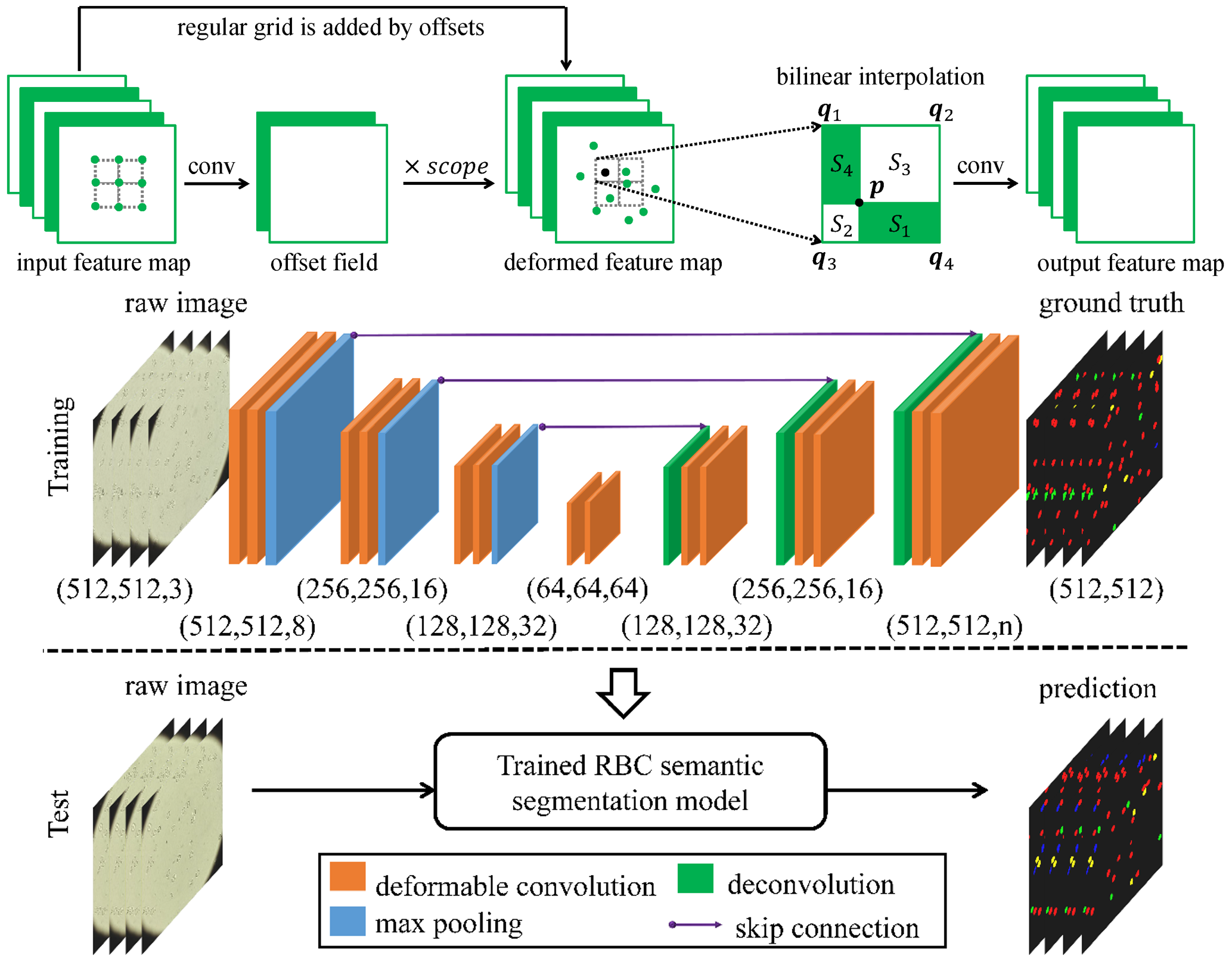
In this work, we adopt a deformable convolution-based deep learning framework to solve the challenge of large variations on the size, shape and viewpoint of the imaging objects. We add deformable convolution layers to the classic U-Net structure and implement the deformable U-Net (dU-Net), which enables free-form deformation of the feature learning process, thus making the network more robust to various cell morphologies and image settings. dU-Net is tested on microscopic red blood cell images from patients with sickle cell disease. Results show that dU-Net can achieve highest accuracy for both binary segmentation and multi-class semantic segmentation tasks, comparing with both unsupervised and state-of-the-art deep learning based supervised segmentation methods. Through detailed investigation of the segmentation results, we further conclude that the performance improvement is mainly caused by the deformable convolution layer, which has better ability to separate the touching cells, discriminate the background noise and predict correct cell shapes without any shape priors.
Repository: https://github.com/XiangLi-Shaun/deformableConvolution_2D
Related publications:
https://ieeexplore.ieee.org/abstract/document/9122550
https://link.springer.com/chapter/10.1007/978-3-030-00937-3_79